It may not be the first time you’ve heard the phrase “size doesn’t matter”, and in this case it could not be much more factual! Although microbes are organisms of tiny proportions, regularly being 1000 times smaller than human somatic cells, our collective microbiome still makes up about 2% of our adult body weight (Warriner et al. 2015). The human microbiome can be accurately described as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body space and have been all but ignored as determinants of health and disease” (Warinner et al. 2015). With recent discoveries of ancient preserved remains, such as the Andean mummy, scientists are able to compare the microbiomes of these ancient humans to the microbiomes of people in present day to gain a greater understanding of the evolution of diet and chronic diseases.
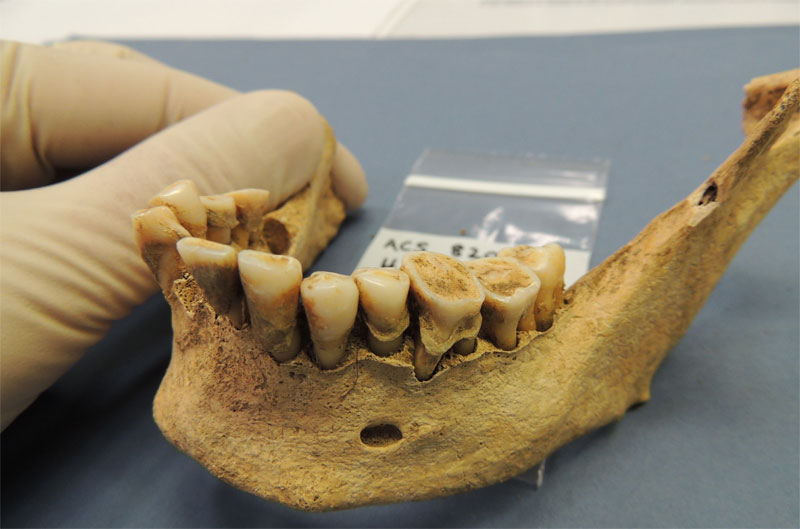
In ancient humans, teeth can be a major source of information due to dental calculus, which is produced when dental plaque becomes mineralized with calcium phosphate, trapping any microbes in the bone-like solid. Because of this fossilization like event, scientists can extract the microbes and study them to find out about changes in the health of humans due to past dietary shifts. When the human diet shifted from a hunter-gatherer diet to one that focussed heavily on carbohydrates during the Neolithic period, oral health began to change as well. By combining skeletal evidence and information determined from the trapped bacteria, scientists have discovered that periodontal disease became much more prevalent during this time period (Rivera-Perez et al. 2015). Along with these oral diseases, a number of chronic diseases are thought to have originated from the decreased microbiome diversity.
As previously mentioned, due to major developments in sanitation, medicine, and industrialization, our relationship with microbes has become immensely different (Warinner et al. 2015). Besides oral diseases, we see a large increase in the percentage of inflammatory diseases such as asthma, allergies, and gum disease in modern day (Levy 2014). Levy adds, “knowing how and why our microbiomes have changed could provide new ways to treating these conditions” (2014). Microbes also give us insight into the resistance of antibiotics (Warinner et al. 2015). Warinner states, “low-level antibiotic resistance has long been present in the human microbiome, illustrating how the oral microbiome functions as both a source and a reservoir of new antibiotic resistance” (2015). Recently, there was an 11th century A.D. Andean mummy discovered which held some very interesting information for scientists. In this unique specimen, scientists found multiple strains of antibiotic resistant pathogens, which is focal in the understanding of evolution of pathogens, and counterintuitively shows resistance to antibiotics may not be inherently attributable to selective pressures from modern medicine (Santiago-Rodriguez 2015). Chronic illness and drug resistance are suggested to be linked to the decreased diversity of microbiomes in the modern human (Warinner et al. 2015). A deeper understanding of ancient microbes will give us the information needed to balance our microbiomes and lessen our susceptibility to many different diseases (Levy 2014).
While humans have made many advances over the past few hundred years involving dietary shifts, improved sanitation, and antibiotic use, these developments have also acted as “weapons of mass microbial disruption” (Warinner et al. 2015). Through analysis of dental calculus, ancient microbiome research shows that these changes in the human way of life has decreased the diversity of the microbiome, leading to many unfortunate diseases including obesity, diabetes and other inflammatory conditions. By acquiring this knowledge of how, when, and why, particular diseases began to occur, scientists are one step closer to finding a way to fight these diseases. Because dental calculus is ubiquitous, more research will continue to take place involving ancient microbes to further expand our knowledge of human biology and diseases.
Levy, S. 2014. Feature: Time Travellers: meet your ancestors’ microbes. New Scientist [Internet]. [cited 2016 Feb 11]; 222(2965): 50-53. Available from: http://eds.a.ebscohost.com/eds/detail/detail?sid=0bd7aa6c-1792-4ff8-85d3-98386ecf06b3sessionmgr4005&vid=2&hid=4113&bdata=JnNpdGU9ZWRzLWxpdmU#AN=S0262407914607880&db=edselp doi: 10.1016/S0262-4079(14)60788-0
Rivera-Perez J, Cano, RJ, Narganes-Storde Y, Chanlatte-Baik, L, Toranzos. 2015. Retroviral DNA Sequences as a means for determining ancient diets [internet]. [cited 2016 March 07]; 10(12): 1-10. Available from: http://eds.a.ebscohost.com/eds/detail/detail?vid=18&sid=8612a147-984b-4da5-a567-a4fd8364dbafsessionmgr4004&hid=4211&bdata=JnNpdGU9ZWRzLWxpdmU#AN=111571193&db=a9h doi: 10.1371/journal.pone.0144951
Santiago-Rodriguez T, Fornaciari G, Luciani S, Dowd S, Toranzos G, Marota I, Cano R. 2015. Gut Microbiome of an 11th Century A.D. Pre-Columbian Andean Mummy. PLoS ONE [Internet]. [cited 2016 March 6]; 1(10): 1-23. Available from: http://eds.b.ebscohost.com.ezproxy.tru.ca/eds/detail/detail?vid=6&sid=fc1393e4-1ed6-402d-b70c-fa196592dab9sessionmgr198&hid=121&bdata=JnNpdGU9ZWRzLWxpdmU#AN=110043439&db=a9h doi: 10.1371/journal.pone.0138135
Warinner C, Speller C, Collins MJ, Lewis CM Jr. 2015. Ancient human microbiomes. J Hum Evol [Internet]. [cited 2016 March 4]; 79(4): 125-136. Available from: http://www.sciencedirect.com.ezproxy.tru.ca/science/article/pii/S0047248414002620